Available Thesis Projects
This is a list of projects for the academic year 2022/2023.
Please contact davide.sassera@unipv.it if you are interested.
When applying for an internship please state which project you apply for, why and what motivations and skills you bring. During the interview you will be questioned regarding the reference articles indicated in each project.
Genomic epidemiology of pathogens (for one Master and Bachelor degree student)
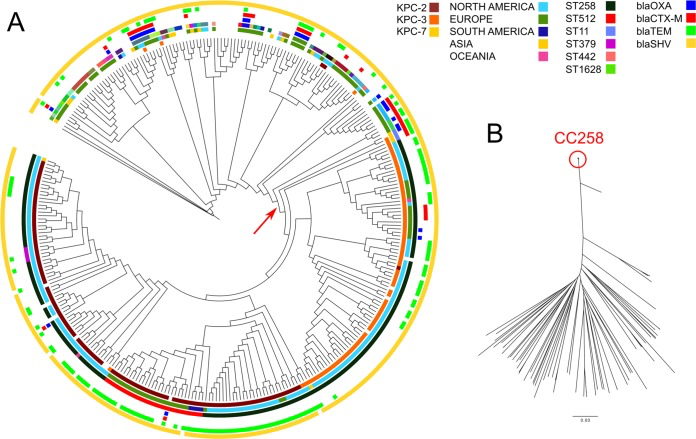
The aim of this project is to analyse by bioinformatic methods the genomes of bacterial and parasitic pathogens, and combine the results with epidemiological and clinical data. This will allow to characterize resistance and virulence traits, to determine epidemiological trajectories and reconstruct outbreaks, but also to reconstruct phylogenies and associate genomic variations to evolutionary trees.
The student will learn the basic techniques of bioinformatic applied to genomic studies. The project is fully bioinformatic with no wet-lab work.
Reference articles: https://pubmed.ncbi.nlm.nih.gov/25367909/ , https://pubmed.ncbi.nlm.nih.gov/31849904/
Comparative genomics of bacterial symbionts (for one Master/Bachelor degree student)
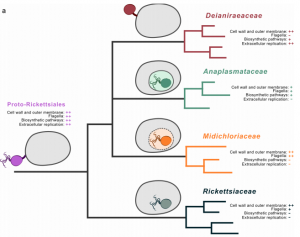 The aim of this project is to sequence and analyse by bioinformatic methods the genomes of novel bacterial symbionts belonging to different lineages (e.g. Rickettsiales, Coxiellaceae, Enterobacteriales) and associated to diverse eukaryotic hosts (e.g. ciliates, ticks, insects). In particular, comparative genomic analyses will be performed in order to elucidate the evolutionary history of the symbionts and the specific adaptations involved in the host-associated lifestyle and interactions
The aim of this project is to sequence and analyse by bioinformatic methods the genomes of novel bacterial symbionts belonging to different lineages (e.g. Rickettsiales, Coxiellaceae, Enterobacteriales) and associated to diverse eukaryotic hosts (e.g. ciliates, ticks, insects). In particular, comparative genomic analyses will be performed in order to elucidate the evolutionary history of the symbionts and the specific adaptations involved in the host-associated lifestyle and interactions
The student will learn the basic techniques of bioinformatic applied to evolutionary studies. The project is fully bioinformatic with no wet-lab work.
Reference article: https://pubmed.ncbi.nlm.nih.gov/31073215/
Molecular screening of vector-borne microorganisms (for one or two Bachelor/Master degree students)
 This project aims to evaluate the presence of pathogens and symbionts in ticks using molecular biology techniques. The students will perform field collection of ticks, DNA extraction, PCR and potentially qPCR, microfluidic PCR, Sanger sequencing. The obtained results will be subjected to phylogenetic and statistical analysis. Previous lab experience is a plus but is not required.
This project aims to evaluate the presence of pathogens and symbionts in ticks using molecular biology techniques. The students will perform field collection of ticks, DNA extraction, PCR and potentially qPCR, microfluidic PCR, Sanger sequencing. The obtained results will be subjected to phylogenetic and statistical analysis. Previous lab experience is a plus but is not required.
Reference article: https://pubmed.ncbi.nlm.nih.gov/33646482/
Metatranscriptomics of ticks (for one Master degree student)
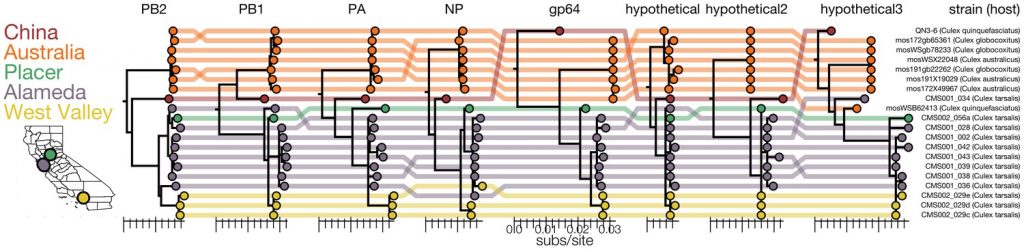
The aim of this project is to develop an innovative approach for characterization of ticks using metatranscriptomics. Such a bioinformatic approach will allow to characterize single ticks from a population genomics and transcription profile point of view in parallel. The student will learn how to manage and analyze transcriptomics data and potentially to perform multi-layered analysis on complex datasets, including machine learning techniques.
Reference article: https://pubmed.ncbi.nlm.nih.gov/33904402/
