Update 26.11.2025
At the moment we do not have any open position, but we are always open for interviews!
Please contact davide.sassera@unipv.it if you are interested.
When applying for an internship please state which project you apply for, why and what motivations and skills you bring. During the interview you will be questioned regarding the reference articles indicated in each project.
Comparative genomics and molecular interactions of bacterial symbionts
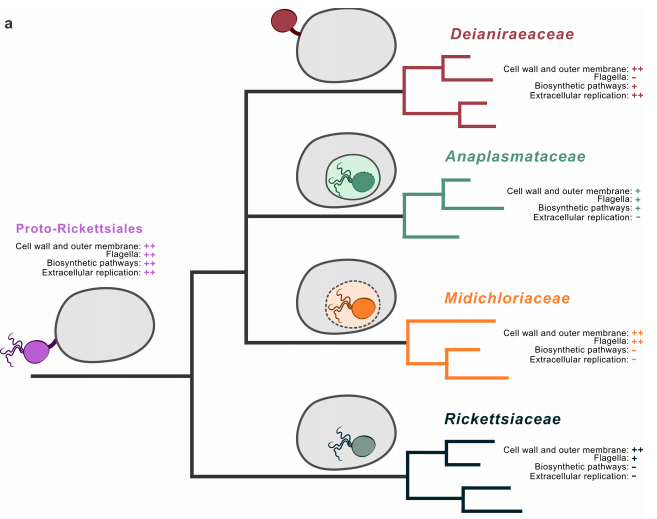
The aim of this project is to sequence and analyze the genomes of bacterial symbionts belonging to the family Rickettsiaceae (including the pathogen Rickettsia spp.) using bioinformatic methods. Comparative genomic analyses will be carried out to elucidate the evolutionary history of these symbionts and to identify the specific adaptations associated with a host-dependent lifestyle. Additionally, structural prediction analyses will be performed to gain a deeper understanding of the molecular interactions between the bacteria and their respective hosts.
Reference articles:
https://pubmed.ncbi.nlm.nih.gov/38173299/
https://pubmed.ncbi.nlm.nih.gov/37305924/
https://pubmed.ncbi.nlm.nih.gov/39488869/
Genomic epidemiology of pathogens
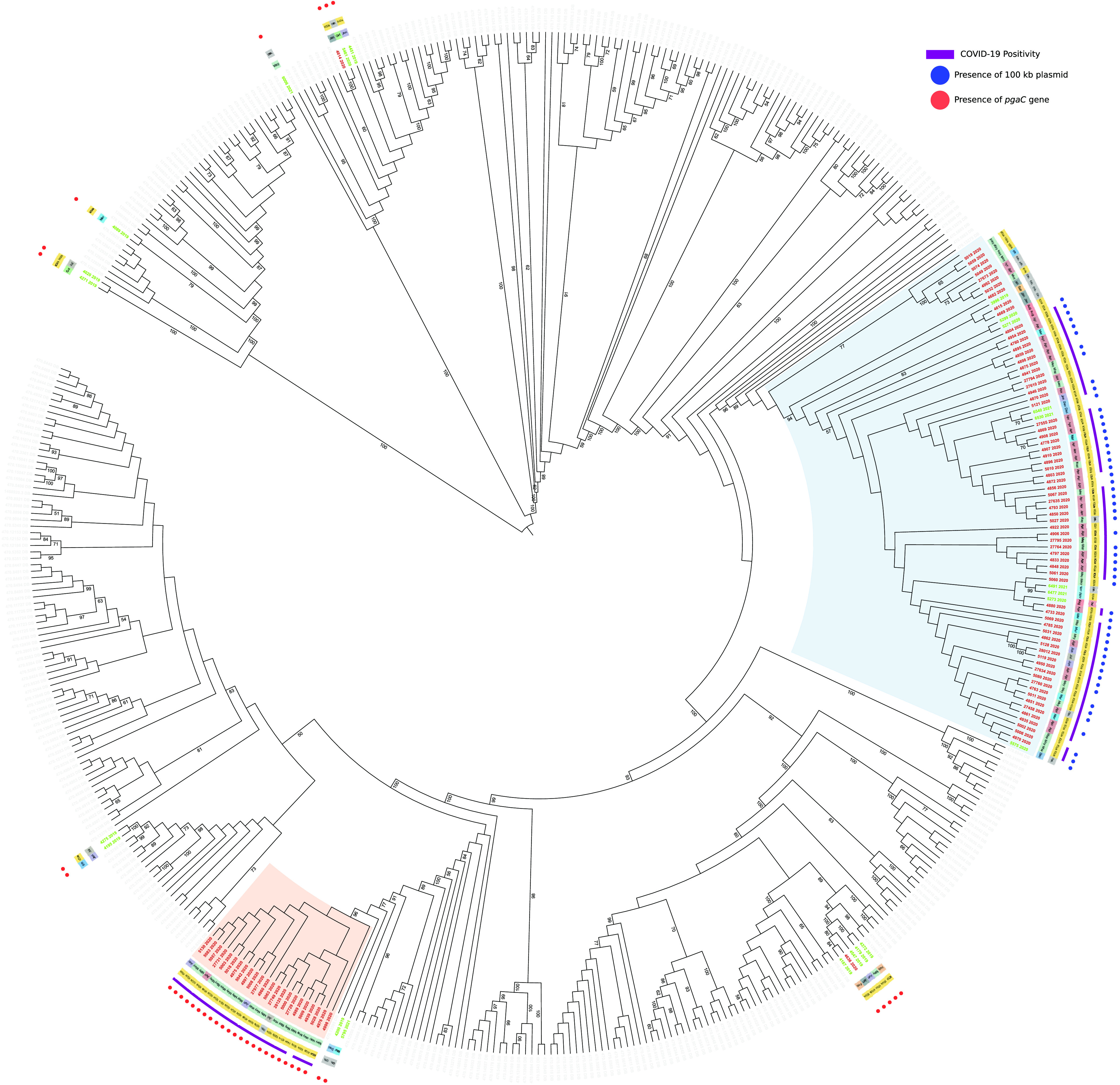
The aim of this project is to analyse by bioinformatic methods the genomes of bacterial and parasitic pathogens, and combine the results with epidemiological and clinical data. This will allow to characterize resistance and virulence traits, to determine epidemiological trajectories and reconstruct outbreaks, but also to reconstruct phylogeny and associate genomic variations to evolutionary trees.
The student will learn the basic techniques of bioinformatic applied to genomic studies. The project is fully bioinformatic with no wet-lab work.
Reference articles:
https://pubmed.ncbi.nlm.nih.gov/31849904/
https://pubmed.ncbi.nlm.nih.gov/38977307/
https://www.biorxiv.org/content/10.1101/2024.11.11.622614v1/
Different sub-project are available:
The project is focused on the genomic epidemiology and evolution of fungal pathogens like Candida spp. using bioinformatic methods, also integrating genomics with clinical data. Main goals will be based on the characterization of resistance and virulence traits, determining epidemiological trajectories and reconstruct possible outbreaks, but also infer phylogenetic reconstruction and associate genomic variations to evolutionary trees. The student will learn the basic techniques of bioinformatic applied to genomic studies. The project is fully bioinformatic with no wet-lab work.
Contact: @DavideSassera, @MichelaVumbaca, @GherardBatistiBiffignandi
The zoonotic parasite Cryptosporidium is a global cause of gastrointestinal disease in humans and animals. We previously investigated the genomic epidemiology of Cryptosporidium parvum in Europe describing the presence of two main populations, one of which causing multiple outbreaks. Future work will aim to study the global evolution of this parasite.
If you want to apply for this position, please carefully read “Bellinzona, Greta et al. “Comparative genomics of Cryptosporidium parvum reveals the emergence of an outbreak-associated population in Europe and its spread to the United States.” Genome research vol. 34,6 877-887. 23 Jul. 2024, doi:10.1101/gr.278830.123”
and contact @DavideSassera and @GretaBellinzona
In silico structural biology to study Host-Symbiont/Pathogen interactions
The goal of this project is to combine omics data with structural biology, particularly AlphaFold-Multimer, to predict interactions between organisms—such as symbionts and their hosts—uncovering molecular processes like immune evasion, metabolic cooperation, and the manipulation of host cell functions.
Current projects involve:
– Interaction between Midichloria mitochondrii and Ixodes ricinus.
– Interaction between Borrelia and its vectors and hosts.
The student will learn the basic techniques of bioinformatic involving -omics analyses and structural biology. The project is fully bioinformatic with no wet-lab work. Previous bioinformatic experience is a plus but is not required.
The project focuses on reconstructing the Borrelia–human interactome. Borrelia, the bacterium responsible for Lyme disease, is known for its ability to evade the immune system and colonize diverse tissues such as the brain, heart, and skin. While a few molecular interactions underlying these processes have been identified, the vast majority remain unknown. By leveraging AlphaFold-Multimer, our goal is to generate the first computationally predicted interactome, uncovering novel protein–protein interactions that may play key roles in Borrelia pathogenesis.
If you want to apply for this position, please carefully read these two papers:
1) Bellinzona, Greta et al. “Accelerating protein-protein interaction screens with reduced AlphaFold-Multimer sampling.” Bioinformatics advances vol. 4,1 vbae153. 11 Oct. 2024, doi:10.1093/bioadv/vbae153
2) Burke, D.F., Bryant, P., Barrio-Hernandez, I. et al. Towards a structurally resolved human protein interaction network. Nat Struct Mol Biol 30, 216–225 (2023). https://doi.org/10.1038/s41594-022-00910-8
Contact: @Davide Sassera, GretaBellinzona
Molecular screening of symbiotic microorganisms in arthropods (Bachelor degree student)

This project aims to evaluate the presence, prevalence and abundance of bacterial symbionts in different lineages of arthropods. To this purpose, molecular biology techniques will be used, in particular, the full-length 16S rRNA gene will be sequenced by long reads to characterize the microbial communities of the host. The obtained results will be analysed by dedicated bioinformatic pipelines. This project will allow future investigations on the identified symbionts, e.g. phylogenetic, genomic and transcriptomic analysis.
This project will involve wet laboratory activities, including DNA extraction, PCR, electrophoresis, sequencing set-up on a variegate set of samples, as well as bioinformatic analyses.
Both Master and Bachelor degree students will perform wet laboratory analysis, Master degree students will take part in data analysis.
Reference article:
https://pubmed.ncbi.nlm.nih.gov/34996376/
https://www.sciencedirect.com/science/article/pii/S2001037019303745#b0010
